RNA Journal Club 8/6/09
CRD-BP Protects the Coding Region of  TrCP1 mRNA from miR-183-Mediated Degradation
TrCP1 mRNA from miR-183-Mediated Degradation
Irina Elcheva, Srikanta Goswami, Felicite K. Noubissi and Vladimir S. Spiegelman
Molecular Cell 35 (2): 240-246, July 31, 2009.
doi:10.1016/j.molcel.2009.06.007
RNA Journal Club 7/30/09
Ars2 Links the Nuclear Cap-Binding Complex to RNA Interference and Cell Proliferation
Joshua J. Gruber, D. Steven Zatechka, Leah R. Sabin, Jeongsik Yong, Julian J. Lum, Mei Kong, Wei-Xing Zong, Zhenxi Zhang, Chi-Kong Lau, Jason Rawlings, Sara Cherry, James N. Ihle, Gideon Dreyfuss and Craig B. Thompson
Cell 138 (2): 328-339, July 24, 2009.
doi:10.1016/j.cell.2009.04.046
This week’s long summary and analysis by David Garcia:
Synopsis:
The authors demonstrate a clear role for the mammalian Ars2 protein in cell proliferation, as well as an interaction with the Cap-Binding Complex. Depletion of Ars2 reduces the miRNA or siRNA directed repression of two reporter constructs, and levels of mature miRNAs for two out of four miRNAs investigated. Ars2 also co-precipitates with Drosha, but not Dicer, and pre- or mature let-7 can rescue some or all of the loss of repression of the reporters associated with Ars2 depletion. The authors are less successful at directly connecting Ars2’s effect on RNAi that they observed to a potential role in primary-miRNA to pre-miRNA processing by Drosha. The processing assays should have tested more substrates, and their variability is not explained clearly enough.
Detail:
This paper aims to demonstrate a critical role for the mammalian protein Ars2 in cell proliferation, and associate this role with other evidence that it also affects the stability or processing efficiency of a couple of primary miRNA transcripts. While initially setting out to study how mammalian Ars2 imparts a resistance to arsenic oxide treatment, the authors discovered previous studies had focused on a partial clone of the protein. In this study, the full-length protein affects arsenic treatment in a manner opposite to what had been demonstrated before; its reduction has a profound influence on cell proliferation; and it contains a number of domains common to RNA binding proteins, and homology to the Arabidopsis protein SERRATE known to affect the processing of primary miRNA transcripts by Dicer-Like 1 (the plant Drosha).
The paper starts by showing that when Ars2 is knocked down using shRNAs, 3T3 cells die more slowly compared to controls when treated with arsenic oxide (Fig1B). Also observed when depleting Ars2 is a cell proliferation defect, using colony formation assays and counting population doublings (Fig1D,F). To investigate further, the authors generate a floxed allele of Ars2 in mESCs, and then generate mice from which they derive immortalized MEFs that they can infect with Cre expressing retrovirus. These Ars2 depleted cells exhibit the same proliferation defects as had been seen with the cell lines (Fig2B). Examination of various tissues from the Ars2 depleted adult mice show that in tissues that have relatively high cell proliferation and normally high expression levels of Ars2, like hematopoietic tissues, there is decreased cellularity or increased apoptosis (Fig2D). In contrast, there are no such differences observed between the transgenic and WT mice in other lower proliferating tissues.
To find out what other proteins Ars2 interacts with, a flagged-Ars2 is expressed, and co-precipitates with several components of the cap-binding complex (CBC), including CBP80, CBP20, and importin-alpha (Fig3A). Ars2 and the CBC interact with 7-Methyl-Guanosine capped RNAs (Fig4A), as would be expected for a component of the CBC. In addition, Ars2 shuttles between the nucleus and cytoplasm (Fig4B), like other CBC components.
Given SERRATE’s known role in miRNA biogenesis in Arabidopsis, the authors address whether the mammalian Ars2 in involved in RNAi. Two types of luciferase reporters are created: one with 3X let-7 (7mer) miRNA binding sites derived from the C. elegans lin-28 3 prime UTR; and another with a perfectly matched site for the let-7 miRNA (~22mer match). In HeLa cells, they first transfect siRNAs to knock down Ars2 or CBP80 or Ago2 (positive control), and then transfect the reporters to see how repression compares to cells treated with a control siRNA. The siRNAs against Ars2 and CBP80 inhibit let-7 directed repression at levels comparable to knockdown of Ago2 for both reporters, with a more pronounced effect on the miRNA sites reporter (Fig5A).
I am surprised knocking down Ago2 only affected repression ~2.5 fold for the perfect site reporter, and not much greater. Let-7 levels are very high in HeLa cells. However, there could be some complications of knocking down Ago2 with an siRNA. Unfortunately, while the authors state a reporter with a mutated let-7 seed site led to a loss of repression under all conditions, they do not show how this loss of repression compares to the loss they observed from Ars2, CBP80, and Ago2 siRNAs. This would have been a useful control to show.
Next the authors show that addition of excess let-7 duplex can rescue loss of Ars2, but not Ago2, for both reporters (Fig5B), suggesting Ars2 may affect some step in miRNA biogenesis before Dicer. Depletion of Ars2 or DGCR8 with two different siRNAs each led to a decrease in mature let-7 levels (~50% reduction)(Fig5C,D), and the same story for mature miR-21 (expressed very highly in HeLa). While they couldn’t detect pre-let-7 by Northern (I’m surprised by this), they did see a reduction in pre-miR-21 in Ars2 depleted HeLa cells. The reduction in pre or mature was not observed for all miRNAs they probed: there was no change for miR-30a or miR-16 (FigS3).
To investigate which step of miRNA processing is affected by Ars2 depletion, they immunoprecipitate a tagged Drosha and Dicer from 293T cells. Ars2 and CBP80 both co-precipitate with Drosha but not Dicer, and this interaction is not RNA dependent (Fig6A). Addition of excess pre-let-7 rescued some of the loss of repression from Ars2 or CBP80 depleted cells (but not Ago2 depleted)(Fig6C). Depletion of Ars2 also led to a decrease in pri-miR-21 levels by qRT-PCR, indicating that Ars2 may also influence the stability of Drosha substrates (Fig6D).
A pri-miRNA processing assay is employed to address the role of Ars2 in affecting the fidelity of Drosha mediated processing. In these assays, they compare the processing of an in vitro transcribed pri-miR-155 between different cell extracts. The authors state that there is an observed reduction in pri to pre processing in the MEF Ars2 KO cells compared to WT (Fig6F). However, it may be problematic to compare two different cell extracts that are almost guaranteed to differ in more than just Ars2 expression. In the Ars2 KO, the “correct” pre band mostly disappears, but a slightly smaller band also present in the WT intensifies, without explanation. They only show data for a single pri-miRNA, 155 (but say comparable results were obtained for miR-21). They quantitate the reduction in “correct” processing as 3-fold for miR-155 (Fig6G).
The paper ends by weakly connecting the observed effect of Ars2 on cell proliferation with its potential effect on miRNA processing. The authors first re-visit the proliferation point by showing that serum starved MEFs that exit the cell cycle lose Ars2 expression (Fig7A). They confirm Ars2’s proliferation dynamic expression in the hematopoietic cell line Bax-/- Bax-/- (Fig7C). A primary miRNA processing assay, comparing processing of pri-miR-155 (in vitro transcribed as before) between 10% serum grown or 0.1% serum starved MEF extracts, is claimed to show a decrease in pri to pre in the Ars2 depleted serum starved extracts (Fig 7D). While there seems to be an increase in heterogeneity in pre, the total reduction in pre in the serum starved cells is not robust. As before, the implication that we are to assume that variability between different cell extracts, from different cell lines, or grown under vastly different conditions, cannot contribute to variability in pri to pre processing is, I think, not well founded. Moreover, the authors only show results from the testing of one miRNA, miR-155. There is insufficient explanation for bands nearby those they deem as “correctly processed.” While the authors have shown clearly how Ars2 affects cell proliferation, interacts with the CBC, and interacts with Drosha, I don’t believe they have clearly explained how Ars2 affects RNAi with their proposed mechanism of affecting miRNA biogenesis. Hopefully there will be stronger data to support this hypothesis in future papers.
The Sounds of Science
While sifting through my iTunes library recently, I came across more than a handful of songs on the subject of science. Sometimes the lyrics directly address science, sometimes the reference is more tangential, and sometimes only the song title is relevant. I’ve included links to audio/video (of mixed quality).
Biology
Do it. Dooooooo it.
This land is mine, this land is free
I’ll do what I want but irresponsibly
It’s evolution, baby
General
The Sounds of Science – The Beastie Boys
I’ve got science for any occasion
Postulating theorems formulating equations
Cheech wizard in a snow blizzard
Eating chicken gizzards with a girl named Lizzy
Dropping science like Galileo dropped the orange
She said I made some new connections to astound them all
in ways we’ve never dreamed about
This is what we all dream about in lab. One of my favorite bands couldn’t have stated it better.
Yeah, damn this thing. Why must it insist on half-a-tick-mark accuracy on my P1000?!
Physics
Why Does The Sun Shine? (The Sun Is A Mass Of Incandescent Gas) – They Might Be Giants
Ok, this song definitely takes the cake for the scienceiest song from a legitimate rock band I’ve ever heard.
You must listen to it–it’s a real hoot.
The sun is a mass of incandescent gas
A gigantic nuclear furnace
Where hydrogen is built into helium
At a temperature of millions of degrees
What Is The Light? – The Flaming Lips
What is the light
That you have
Shining all around you?
Is it chemically derived?
Colors are a fascinating physical phenomenon. This song has a real nice melody, and sounds really sciencey too.
Chemistry
Polyethylene (Parts 1&2) – Radiohead
So sell your suit and tie and come and live with me
Leukemia schizophrenia polyethylene
There is no significant risk to your health
She used to be beautiful once as wellPlastic bag, middle class, polyethylene
Decaffeinate, unleaded, keep all surfaces clean
The Chemistry Of Common Life – Fucked Up
Here but for the spinning of a sphere,
Electric skies and vibrations rise the breach,
the birth, the seed inside,
The chemistry of common life
Chemical Elements
Cobain was speaking about atomic number 3; or maybe he was talking about mood stabilizing drugs.
Who cares the song rocks.
The great Detroit troubadour Jack White went simple for the lyrics to this song.
Oh yeah, and this track is off their album White Blood Cells.
AAaahhhhhhhhhhhhh!
AAaahhhhhhhhhhhhh!
AAaahhhhhhhhhhhhh!
BioMedicine
Better Living Through Chemistry – Queens Of The Stone Age
The blue pill opens your eyes
Is there a better way?
A new religion prescribed
To those without the faith
Sister Morphine – The Rolling Stones
Here I lie in my hospital bed
Tell me, Sister Morphine, when are you coming round again?
Oh, I don’t think I can wait that long
Oh, you see that I’m not that strong
Hey baby you better come here quick
This ol’ cocaine is making me sick
Cocaine all around my brain
Yeah, it’ll do that.
Space
If you believed they put a man on the moon, man on the moon
. . . Newton got beaned by the apple good. Yeah, yeah, yeah, yeah
. . . Mister Charles Darwin had the gall to ask. Yeah, yeah, yeah, yeah
The Apollo Programme Was A Hoax – Refused
No it wasn’t. It happened. Listen to the R.E.M. song.
Polemic and Beyond
Science fails to recognize the single most
potent element of human existence
letting the reigns go to the unfolding
is faith, faith, faith, faith
What Would Wolves Do? – Les Savy Fav
The world may seem cruel
The world may hate us
In time we will show the world why the world made us
God Makes No Mistakes – Loretta Lynn
You might be wondering why a song with this title is on this list. I find the lyrics really fascinating actually; while a scientist’s explanation for these “mistakes” may be far simpler, they are probably unlikely to resonate as profoundly as Lynn’s with most people.
Why I’ve heard people say
Why is my child blind
Why is that old drunk still livin’
When a daddy like mine is dyin’
our blessed father gives us life
has the power to take it away
There’s no reason for what he does
God makes no mistakes
RNA Journal Club 7/23/09
Cellular MicroRNA and P Bodies Modulate Host-HIV-1 Interactions
Robin Nathans, Chia-ying Chu, Anna Kristina Serquina, Chih-Chung Lu, Hong Cao, and Tariq M. Rana
Molecular Cell 34 (6): 696–709, June 2009.
doi:10.1016/j.molcel.2009.06.003
This week’s summary and scrupulous analysis by Anonymous:
The authors showed that a host microRNA (miRNA), mir-29a, targets the 3’ untranslated region (3’ UTR) of HIV-1 directly and negatively regulates viral expression. mir-29a has two other family members (with the same seed sequence) that are also able to repress viral expression. In addition, the paper suggested that this negative regulation contributes to the latency of HIV-1 via the following model: Upon integration into the host genome and transcription, viral mRNAs are transported out into the cytoplasm where translation of viral proteins and virus assembly usually takes place. If however, the HIV-1 3’ UTR is targeted by mir-29a, the viral mRNA is sequestered into P bodies, translation is suppressed and virus assembly is prevented. Activation could occur when viral mRNAs are released from P bodies after certain stimuli. If this model is true, then mir-29a-mediated regulation would be acting as a checkpoint from viral latency to activation.
While the authors convincingly demonstrated that mir-29a negatively regulates viral expression via a direct interaction with the HIV-1 3’ UTR, the evidence presented for P body sequestration is not as strong. The viral RNA is certainly sequestered into cellular foci but three of the four markers used to identify P bodies in the paper can also be found in stress granules. The authors made a point that the negative regulation is due to translational suppression and not mRNA destabilization but not much was done to probe that the latter was not taking place.
Still, the authors did a nice job tying the relevance of this miRNA-mediated regulation to viral latency. Aligning the 3’ UTRs of various HIV-1 subtypes, they found that the mir-29a target site in the 3’ UTR is highly conserved. This is true for most subtypes except the O group HIV-1 RNAs, in which the non-conserved nucleotides in the seed match region would abolish interaction with mir-29a. The authors highlight the fact that O group HIV-1 is endemic to certain parts of Africa and is typically 100-fold less infectious than the widely-circulating M group HIV-1, whose viral subtypes maintain the conserved nucleotides in the mir-29a seed match site. Hence it is plausible that mir-29a interaction and HIV-1 infection capability could be linked. If this is true, it is remarkable that the relatively recently-evolved HIV-1 is able to co-opt the more ancient mir-29a (conserved across humans, mice, rats, dogs and chickens) to modulate its own life cycle.
It is worth mentioning that while HIV-1 produces more than 30 mRNAs, almost all of them share the same 3’ UTR. Hence even a single miRNA-mediated downregulation would prevent other viral proteins, such as the viral transcription activator Tat, from being made. This, in turn, would reinforce latency. Having said that, mir-29a is not the first miRNA identified to target the 3’ UTR of HIV-1. A previous paper (ref.) found that several miRNAs (though mir-29a was not one of them) target the HIV-1 3’ UTR and contributes to latency in resting primary CD4+ T lymphocytes, which are the cells usually infected by HIV-1. In the subtype alignment described above, the 3’ UTR is well-conserved even beyond the mir-29a seed match site. It would be interesting to see if the seed matches of these miRNAs also coincide with these other highly-conserved segments.
Reference:
Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes
Huang et al., (2007) Nature Medicine 13: 1241-1247.
You’d Prefer An Argonaute Banned in China

I have just received word from a close colleague of mine traveling in China that this blog is blocked there.
Update:
Actually, apparently all WordPress blogs are blocked in China. Thanks for the tip Yang. (See comments below.) Damn, and I could be getting like 5 million more views per day.
Evolution is Awesome
A new article in the August issue of National Geographic gives me another reason to embrace natural selection–um, despite my many fitness shortcomings. The Art of Deception describes some survival trickery employed by various inhabitants of the Panamanian rainforest and elsewhere, and being in National Geographic, there are a bunch of nice pictures with educational captions.
My favorites: Cathedra serrata, the insect with alarmingly large, fake insect eyes–because it’s always good to have a backup plan; and the nematode that, visually speaking, turns your garden variety Cephalotes atratus into a delicious cherry-ant, an irresistible Central American rainforest hors d’oeuvre.
The Art of Deception, National Geographic, pgs. 70-87, August 2009. Words by Natalie Angier, Photographs by Christian Ziegler.
RNA Journal Club 7/16/09
Therapeutic microRNA Delivery Suppresses Tumorigenesis in a Murine Liver Cancer Model
Janaiah Kota, Raghu R. Chivukula, Kathryn A. O’Donnell, Erik A. Wentzel, Chrystal L. Montgomery, Hun-Way Hwang, Tsung-Cheng Chang, Perumal Vivekanandan, Michael Torbenson, K. Reed Clark, Jerry R. Mendell and Joshua T. Mendell
Cell 137 (6): 1005-1017, June 2009.
doi:10.1016/j.cell.2009.04.021
This week’s exemplary summary and analysis by Anonymous:
A recent report by Kota and colleagues describes the systemic delivery of a tumor suppressive miRNA, miR-26a, to a murine model of hepatocellular carcinoma (HCC). In this study, it was shown that miR-26a, a miRNA previously shown to be down-regulated by the c-Myc oncogene, functions as a tumor suppressor via induction of a G1 arrest in a human HCC cell line. This G1 arrest was ascribed to repression of miR-26 targets Cyclin D2 and Cyclin E2. Furthermore, they described an adeno-associated viral vector that would express miR-26a in combination with eGFP. This virus was then delivered to mice over-expressing c-Myc in the liver, which has previously been shown to induce liver tumors. Compared to tumor-bearing mice infected with a virus expressing only eGFP, tumor-bearing mice infected with the virus expressing both eGFP and miR-26a had a substantial reduction in tumor burden without gross toxicity to the liver and other organs. This provides the first example of systemic miRNA delivery suppressing an established tumor in vivo.
While this observation has important therapeutic implications, there are several limitations to the study. First, while the in vitro data suggests that miR-26a acts as a tumor suppressor via cell cycle arrest, the in vivo data for miR-26a causing cell cycle exit is unconvincing. In particular, the use of Ki67 intensity as a measure of cell cycle status is not legitimate, as Ki67 is either present in cycling cells or absent in non-cycling cells. Thus, it appears that the more likely mechanism of tumor suppression in vivo is apoptosis, due to the robust increase in TUNEL staining. However, it is not clear which targets of miR-26a contribute to increased apoptosis. Moreover, while this study proves that miR-26a can suppress early-stage c-Myc liver tumors, it is not clear whether miR-26a can suppress late-stage, invasive HCC. Since most patients with HCC show up with significantly advanced disease, examination of miR-26a delivery in more aggressive lesions would be an important next step.
RNA Journal Club 7/9/09
Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs
Aly A Khan, Doron Betel, Martin L Miller, Chris Sander, Christina S Leslie & Debora S Marks
Nature Biotechnology 27 (6): 549-55, June 2009.
doi:10.1038/nbt.1543
This week’s cerebral analysis by Graeme Doran:
Investigating the application of small RNAs to destabilize specific mRNA targets, researchers have observed a variety of non-specific ‘off-target’ effects – alterations in the expression level of mRNAs that do not contain a perfectly complementary target site for the small interfering RNA transfected into cells. In a minority of cases, the target mRNA is actually stabilized!
Briefly, ‘off-target’ effects have been broadly understood to derive from 4 main sources:
a) Partial complementarity between the small interfering RNA and mRNAs in the cell.
b) Stress responses due to transfection of foreign RNA.
c) Downstream secondary gene expression changes due to silencing of a specific target.
d) Disruption of the endogenous miRNA regulatory network due to competition between the cellular miRNA machinery and exogenously supplied small interfering or short hairpin RNAs.
To date, conflicting experiments have suggested that some si/shRNAs may inhibit endogenous miRNA activity in some scenarios, particularly when the silencing RNAs are present at high levels. This may occur through saturation of either or both the RISC and DICER activities in the cell, depending on the type of small RNA used.
This study from Khan et al. attempts a broad survey of previously published small RNA transfection experiments.
The key evidence presented are:
1) Transfection of small RNAs into immortalised cell lines produces a consistent de-repression of genes that contain an endogenous miRNA target site within their 3’UTR.
2) The extent of de-repression is quantitative – that is it depends both upon the extent of endogenous miRNA repression on a specific UTR, and the concentration of siRNA used in the transfection.
3) Cellular ARGONAUTE/RISC activity is likely subject to loading competition between small RNAs within cells.
Compiling an extensive set of data leads to some brevity of description in the methods. The authors use 4-species conservation as a criterion for miRNA site prediction. One would expect that the observed de-repression effect would be independent of the seed site conservation, as it is commonly possible to see miRNA repression on non-conserved seed sites, and the authors do not make clear whether this signal was present for the non-conserved site data or not. Further, the normalization of predicted target site sets is not well described. One might envisage that the ‘baseline’ gene sets with no endogenous or exogenous seed sites would be shorter on average than gene sets with 1 or more conserved sites. Shorter UTRs have less scope for regulation by miRNAs or other
RNA binding factors, and so maybe would have less regulation to perturb.
The core data (Figure 2) indicates a consistent and significant de-repression of UTRs that contain conserved (higher confidence) endogenous miRNA target sites, but the data is correlative rather than conclusive – and would have been greatly enhanced by a simple luciferase assay to precisely determine the contribution of individual sequences to repression/de-repression in the context described. As is, the model and the data fit each other, but other modes of de-repression are not discounted experimentally. Data suggesting that the de-repression is dose-dependent seem to rely on relatively small sets of genes selected specifically for responsiveness to siRNA treatment and thus this aspect of the story is less convincing. Furthermore, there is little proof that concentrations required by ‘good’ siRNAs to silence genes (1-10nM) have a significant de-repressive effect on miRNA targets, and so the importance of the observed effect in therapeutic situations where delivery concentrations are low is still an open question.
The core findings of this paper, and the methodology employed, open up some interesting questions about the basic biology of RISC competition in cells. How, for instance, do rapidly induced miRNAs (such as miR-21 upon SMAD pathway activation) interact with the endogenous pool of miRISC? Is there present a surplus of free ARGONAUTE for such instances, or is ARGONAUTE protein concentration limiting in the cell? Do endogenous siRNAs – generated on cell stress or viral infection – displace miRNAs from their silencing role, and what is the half-life of endo-siRNA loaded RISC in vivo? Tumors commonly downregulate miRNA activity, and DICER depletion enhances tumorigenesis. Can this also be achieved by limiting the available miRNA RISC in tumor cells?
Broadly speaking this was a stimulating paper, and whilst I don’t think that it breaks new ground in considering ‘off-target’ effects of small RNAs within cells, it provides some of the most convincing evidence of miRNA target de-repression to date. And beyond this, it provides the thought provoking concept of RISC competition, new questions upon which to focus, and a validated method for analyzing miRNA target de-repression.
Hawt RNA Blogs
Yikes.
As YPAA seeks to spread its scientific cred throughout the blogosphere, I am keen on knowing what other RNA blogs I will step over. So what’s the other hot RNA blog right now? Probably this one: The Romantic Novelists’ Association Blog.
Their name is much more descriptive than YPAA; the topic is far more sensorial. My blog has recently spotlighted literature with titles like, Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. The other RNA blog, No Longer Forbidden and Surrender to the Playboy Sheikh, among others. A recent update of theirs has a recipe for ice cream, which apparently goes really well with romance novels.
So I ask, what goes really well with RNA primary literature?
Pizza. Pizza is the answer.

mediabistro.com

spicemagazineonline.com
RNA Journal Club 7/2/09
MicroRNA-mediated switching of chromatin-remodelling complexes in neural development
Andrew S. Yoo, Brett T. Staahl, Lei Chen & Gerald R. Crabtree
Nature AOP June 29, 2009.
Nature 460 (7255): 642-646, July 30, 2009.
doi:10.1038/nature08139
The History of the Argonaute, Part 1
Why was the the Argonaute (Ago) protein named “Argonaute?” Why wasn’t it named “The Slicenator”, or “Chopinator” or some other cool sounding name reflecting the activity many Ago proteins possess? Or, following Eric Lander‘s description of Ago’s activity as “enzymatic kung-fu” (beginning at 7:20 in Part 1) in the wonderfully done NOVA program introducing RNAi below, how about “sensei”?
Part 1 of the NOVA RNAi program:
Part 2 of the NOVA RNAi program:
The answer to the above question is that the cleavage activity of plant AGO1 of nearly perfect matched targets was not completely worked out at the time it was named. What follows below is a simple graphic depicting the most well characterized domains of Argonaute, a very brief history of how the protein was named, and where the name comes from.

The above graphic is from a class presentation I gave in 2007. Not shown/labeled are the N-terminal or Mid domains also common to eukaryotic Argonaute proteins.

Arabodopsis AGO1 mutant, 1998 (1)

Drawing of Argonauta Argo, male and female, 18th century (2)
The name Argonaute comes from phenotypes observed for AGO1 mutants in Arabidopsis thaliana by Bohmert and collaborators in 1998 (1). To the researchers eyes, the plants resembled the tentacles of the pelagic octopus, Argonauta argo. In the paper, the authors state:
Because of their unusual appearance, which reminded us of a small squid, we named these mutants argonaute.
So then how were the marine Argonauts named? It is thought that early taxonomists were enamored with tales of sitings of Argonauts “sailing” along the surface of the sea; females using their paper-thin eggcase shell as a boat, and their webbed dorsal arms (in the drawing above the webbed portions are resting against side of shell) raised above the surface acting as sails (6). (This method of propulsion is today considered a myth, as it has never been observed by marine scientists.) To these taxonomists perhaps this image bore semblance to great ancient wooden sail ships, like “Argo”, (built by Argus, who may have come from the city of Argos), sailed by Jason and the Argonauts in Greek mythology. The Argonauts –named for the ship they sailed– were a group of brawny, fearless men selected by Jason to join him on his perilous journey to recover the “Golden Fleece”, an undertaking that would finally allow him to rightfully claim the throne as king of Iolcus.
In 1963, a movie was released depicting this mythical tale. The movie is pretty good, and apparently was a special effects pioneer in its day. An unofficial trailer:
The word Argonaute has been used in a variety of other contexts as well. For example, in the 1950’s the French navy named their flagship submarine “Argonaute.” The “Argonaut Conference” was the codename for the Yalta Conference held in Crimea in 1945 that brought together Winston Churchill, FDR, and Joseph Stalin.

In attendance at the Argonaut Conference, 1945: Winston Churchill, FDR, and Joseph Stalin (3)
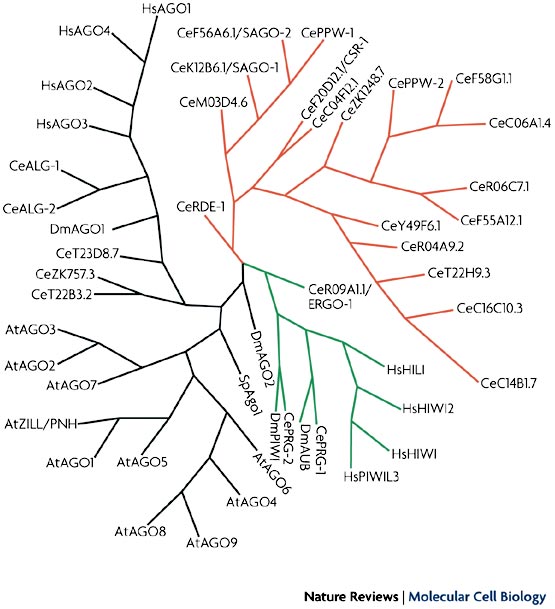
Argonaute phylogenetic tree, present day (5). Argonaute-like group in black, PIWI-like group in green, C. elegans group 3 in red
A sharp increase in genome sequencing and small RNA research in the last decade has lead to the discovery of many more Argonaute genes (including Ago, PIWI-like, and Group 3 in worms (4)), a trend that will surely continue.
Presently in C. elegans, there are >25 known Argonaute genes; 10 in plants; 8 in humans (4 Agos and 4 PIWIs) (5).
Feeling a bit depleted of Argonautes myself, I reckon I’d prefer a few more.
References:
- Bohmert et al., The EMBO Journal Vol.17 No.1 pp.170–180, 1998.
- Internet (reference forthcoming)
- http://en.wikipedia.org/wiki/Yalta_Conference
- Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi.; Cell. 2006
- Hutvagner and Simard, Nature Reviews Molecular Cell Biology 9, 22-32, January 2008.
- http://researchdata.museum.vic.gov.au/argosearch/index.html
RNA Journal Club 6/25/09
Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps
Sung Wook Chi, Julie B. Zang, Aldo Mele & Robert B. Darnell
Nature, Advance Online Publication, June 17 2009.
Nature 460 (7254): 479-486, July 23 2009.
doi:10.1038/nature08170
This week’s deep summary and analysis by Noah Spies:
Despite the best efforts of numerous labs over the last decade, studying microRNA—messenger RNA interactions is still a slow and error-prone process of computational predictions based around sequence conservation (and a host of other sequence elements), supported by luciferase reporter assays, microRNA-transfection followed by micro-array or mass spec analysis, knockouts of microRNAs and components of their pathway, and other methods. So, it is with great excitement and some frustration that we receive this HITS-CLIP paper from the Darnell lab.
In a late 2008 paper, Darnell and colleagues developed the “HITS-CLIP” method, short for high-throughput sequencing of RNA isolated by cross-linking immunoprecipitation. This mouthful-of-an-acronym method involves using ultraviolet light to cross-link proteins to nucleic acids, allowing stringent immunoprecipitation of direct protein—RNA complexes. In Chi et al (2009), the Darnell lab has focused this method on identifying interactions between the workhorse of the microRNA-induced silencing complex, Argonaute (Ago), the microRNAs bound in Ago, and the mRNAs targeted by those microRNAs. The authors found that immunoprecipitation of Ago from cross-linked cells produced two populations of Ago-RNA complexes: (1) Ago—microRNA complexes, which run at about 110 kDa after partial RNase digestion, and (2) Ago-mRNA complexes, which run closer to 130 kDa. By isolating these RNA populations separately and sequencing using Illumina, the authors were able to globally identify both Ago—microRNA and Ago-mRNA interactions.
The difficulty in analyzing these data comes from the heterogeneous population of microRNAs: which Ago-mRNA sequence tag corresponds to an Ago with which microRNA loaded? The authors first use a clever approach they dub “in silico CLIP” to simulate distributing sequence tags across messenger RNAs based on mRNA expression. This simulation provides a background level for the number of tags that would be expected by chance to simultaneously overlap one another, forming clusters. The authors then identify significantly enriched mRNA-sequencing tag clusters, and show that tags in most of these clusters are tightly distributed around the center, giving a sharp peak. For each cluster, then, the authors can search for 6—8mer microRNA seed matches within the cluster, and suggest which microRNA bound which mRNA clusters.
There was a significant enrichment of clusters at both ends of 3′ untranslated regions (UTRs), as was expected given prior research that most functional microRNA targets are in these regions. This study also identified many Ago—mRNA clusters in coding sequence, although not above background, and in introns and intergenic sequence, though the authors did not explore the explanation that these may simply be the result of unannotated transcripts and retained introns.
This method has the advantage over computational predictions of identifying true Ago—mRNA interactions, but these interactions do not necessarily result in noticeable down-regulation of the messenger RNA. To begin to assess how often these Ago—mRNA interactions are productive, the authors transfected a brain-specific microRNA, miR-124, into the cervical cancer cell line HeLa, and then used HITS-CLIP to identify Ago—mRNA clusters. The authors found that those mRNAs apparently bound by miR-124, according to HITS-CLIP, were significantly downregulated following transfection when compared to those with miR-124 sites computationally predicted by TargetScan. This was true at both the protein and the mRNA level across all transcripts, as well as when only looking at the brain-expressed messenger RNAs at the mRNA level, although brain-expressed genes did not show a convincing down-regulation at the protein level.
The authors end with a faulty Gene-Ontology—based analysis, comparing HITS-CLIP to previously published microRNA-target predictions. For the most highly expressed microRNAs in their study, the authors analyzed enrichment of various GO categories in HITS-CLIP mRNA clusters with the associated seed site. They found significant enrichment for several of these microRNAs for several of these neuronal GO categories. In comparing these results to microRNA-target predictions, the authors compared mRNAs with and without predicted microRNA target sites. However, this ignores the fact that many genes have very little conservation in their 3′ UTRs, and hence could not be predicted as targets of any microRNA. A better comparison might take transcripts targeted by non-expressed microRNAs as the background set, and compare these to those predicted to be targeted by the highly expressed microRNAs.
In summary, this is an exciting and powerful new technique, which will quickly broaden our understanding of microRNA regulation. A few issues marred what could have been an exceptionally interesting paper. First, the authors seemingly randomly cherry-pick their data for each figure panel, sometimes choosing conserved microRNAs, sometimes non-conserved; sometimes those clusters present in all their replicates, sometimes only those in two or more replicates; sometimes the top 30 most-expressed microRNAs, sometimes only the top 20. These decisions may have been well founded, or the results were similar regardless of which data they chose, but without clear explanations of why they conducted their analyses the way they did, it is difficult to express confidence in the robustness of their results. Secondly, the HITS-CLIP method has a huge advantage over target prediction methods in being able to identify non-conserved target sites, and yet the authors restricted most of their analyses to only those conserved microRNA targets. Finally, the authors chose not to make the raw HITS-CLIP sequencing data readily available online (submission to the NCBI Short Read Archive is the standard for sequencing data, as GEO is the standard for micro-array data), although one can hope that this will be rectified in the near future.
Update 7/24/09:
As Dr. Darnell calls attention to in his comment below, all of the raw data and UCSC links are now available. For them, visit the Darnell Lab Ago HITS-CLIP website here.
Modest Milestone
Well it took a little over three months, but YPAA has just attained it’s first one thousand total views. Hooray.
Thanks for visiting.



leave a comment